Research lines
Designer gene regulation: disclosing design principles and programming new functions in living cells.
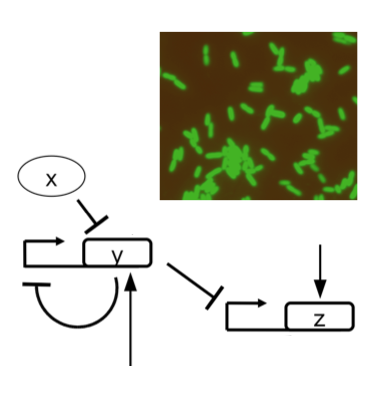
We engineer synthetic gene circuits aimed at (re-)programming living cells with repurposed and de novo designed regulatory elements. We are especially appealed by post-transcriptional control mechanisms. We use bacteria as model organisms for foundational systems and synthetic biology. We also focus on plants as organisms with environmental impact. In addition, we develop computational models at different levels of intracellular organization (molecular, genetic, and genomic) to understand in a quantitative way the bases of gene regulation. Of particular interest is the study of how cells manage the stochasticity underlying gene expression programs
Diagnostics and nanobiocomputing: detecting infectious agents and genetic biomarkers with engineered molecular systems.
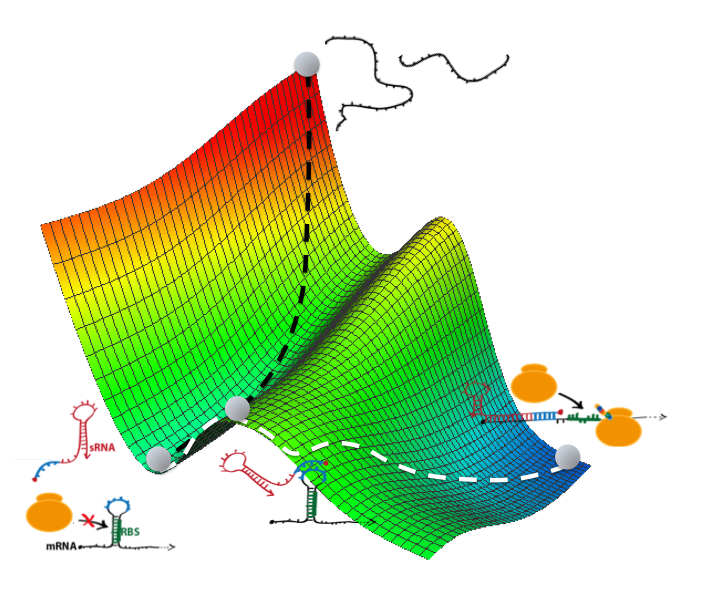
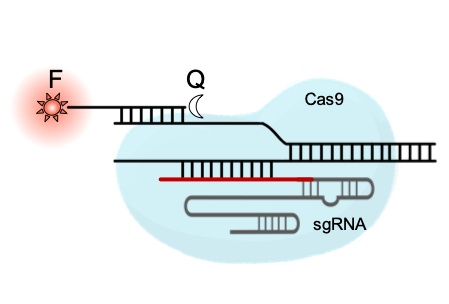
We engineer biomolecular systems able to recognize specific nucleic acids. We mainly focus on CRISPR-based detection methods, exploting different nucleases. In addition, we explore the use of cell-free expression systems and stimulus-responsive nanomaterials to develop novel strategies for molecular computing, with the aim of processing molecular information with precision in vitro. We apply these systems to detect viruses, bacteria, and genetic (e.g., cancer) biomarkers from clinical and environmental samples.
Engineering and evolution of molecular automata: re-programming naturally occurring self-assembled and self-operating nanobiostructures.
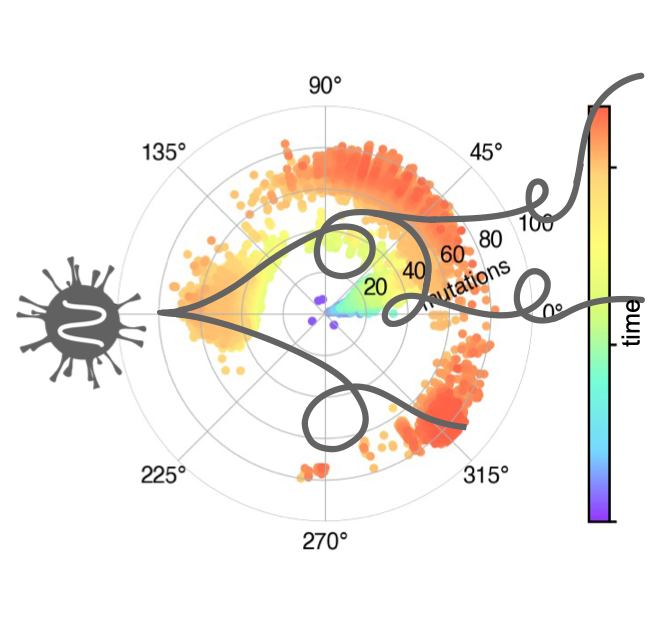
We engineer genetic cassettes encoding molecular automata with potential therapeutic uses and industrial applications. We mainly focus on virus-based nanoparticles, from bacteriophages and plant viruses. In addition, we study how genetically encoded molecular automata evolve. We follow a data-driven approach from whole-genome sequencing to study the mutation accumulation process in viruses and other biological entities that behave like automata using time-dependent probabilistic mathematical models that describe (non-)Brownian motions, as well as using non-equilibrium thermodynamics.
Current funding
Spanish Ministry of Science (Prueba de Concepto, 2022-2024).
Spanish Ministry of Science (I+D Conocimiento, 2022-2025).
Generalitat Valenciana (Expresiones Interes Covid-19, 2022-2023).
CSIC-EU (PTI Salud Global, NextGenerationEU, 2021-2023).
Former funding
Generalitat Valenciana (Junior Investigator, 2020-2022).
European Commission (H2020-MSCA-ITN, 2019-2022).
University of Valencia (VLC-Biomed, 2021).
CSIC (Cuenta la Ciencia, 2021).
CRUE-Banco Santander (Supera Covid-19, 2020-2021).
Spanish Ministry of Science (I+D Conocimiento, 2019-2021).
Spanish Ministry of Economy (I+D Excelencia, 2016-2018).
Generalitat Valenciana (Grupos Emergentes, 2016-2017).
BBVA Foundation (Leonardo, 2015-2016).
Spanish Ministry of Economy (Explora, 2015-2016).
CSIC (Intramural Inicio, 2014-2015).







 BioSystems Design @ Rodrigolab
BioSystems Design @ Rodrigolab